Nadam-accelerated Scalable and Robust CP Decomposition
Introduction
Tensor decompositions is a generalization of matrix decompostions in three or higher-dimensional space. One of the most widely used model is the canonical polyadic (CP) form, also known as parallel factor analysis (PARAFAC) or canonical decomposition (CANDECOMP). Unlike applying the independent component analysis (ICA) or the principal component analysis (PCA) on the matricized / unfolded data for brain network identification, the CP decomposition can capture the low rank struture inherit in the higher-dimensional data. Moreover, it does not need to impose any constraint (in order to have a unique solution), such as independence in ICA or orthogonality in PCA, which may not be realistic as brain networks can overlap and be correlated in both space and time.
Nadam-Accelerated SCAlable and Robust CP Decomposition (NASCAR) is a pipeline for brain network identification across multiple subjects from asynchronous function magnetic resonance imaging (fMRI) data. We first used BrainSync (Joshi et al. 2018) to align fMRI time-series across subjects. As with our earlier work, we used a Scalable and Robust Sequential Canonical Polyadic Decomposition (SRSCPD) framework, where lower rank solutions were used as a warm-start initialization. Inside SRSCPD, we applied Nesterov-accelerated adaptive moment estimation (Nadam) (Dozat, 2016) to a full-gradient search to simultaneously estimate all components of the CP model, to further improve its robustness and scalability.
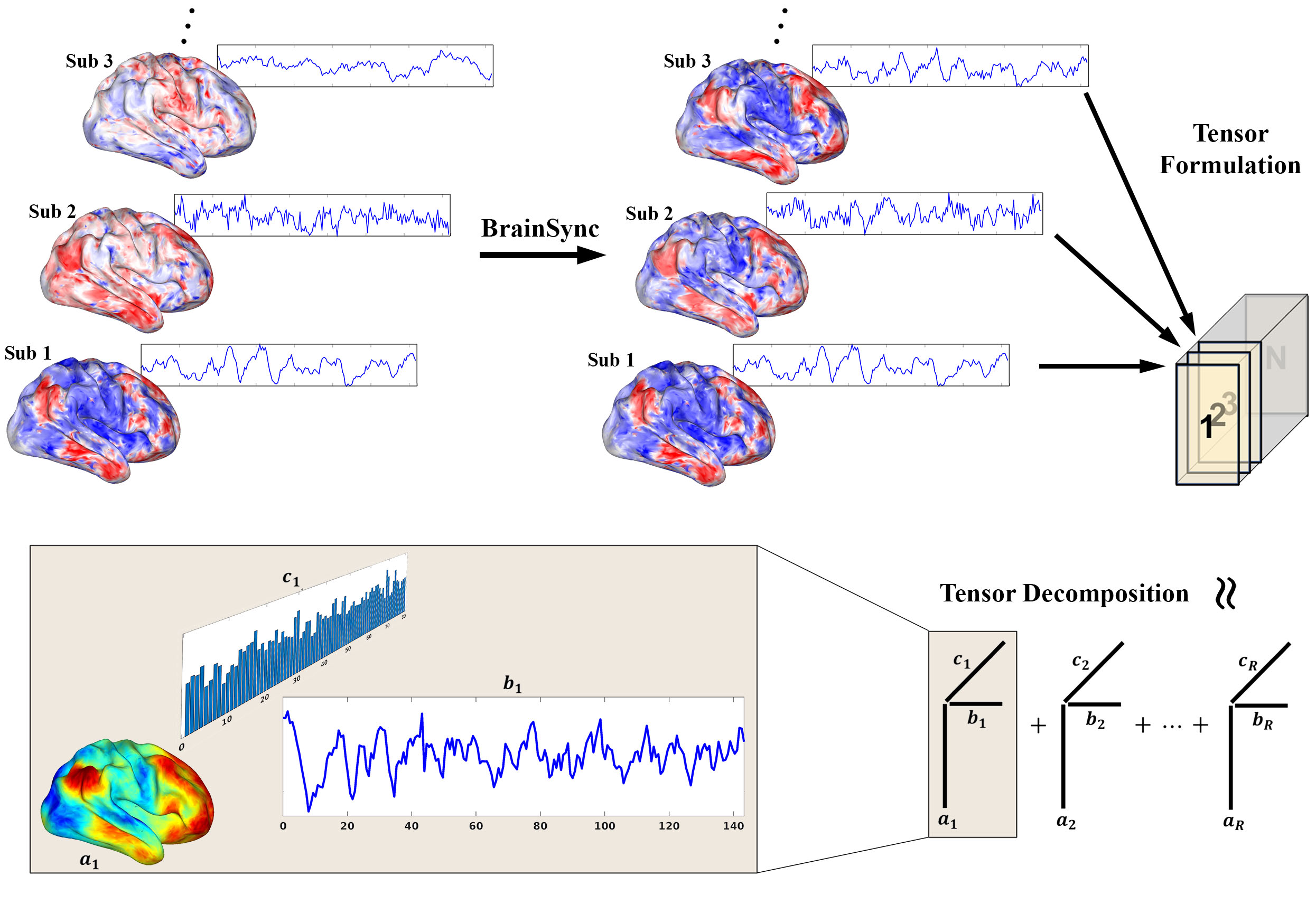
This page provides a MATLAB implementation of NASCAR described in
J. Li, Y. Liu, J. L. Wisnowski, R. M. Leahy, “Identification of overlapping and interacting networks reveals intrinsic spatiotemporal organization of the human brain”, NeuroImage, vol. 270, p. 119944, 2023.
J. Li, J. L. Wisnowski, A. A. Joshi, R. M. Leahy, “Robust brain network identification from multi-subject asynchronous fMRI data”, NeuroImage, vol. 227, p. 117615, 2021.
J. Li, J. L. Wisnowski, A. A. Joshi, R. M. Leahy, “Brain network identification in asynchronous task fMRI data using robust and scalable tensor decomposition”, Proc. SPIE Medical Imaging 2019: Image Processing, San Diego, CA, Mar. 2019, pp. 164–172.
Please cite the papers above in your publications if you have used NASCAR in your research.
Disclaimer
IN NO EVENT SHALL THE AUTHORS, THE CONTRIBUTORS, THE DISTRIBUTORS, THE UNIVERSITY OF SOUTHERN CALIFORNIA AND THE MASSACHUSETTS GENERAL HOSPITAL (“AUTHORS”) BE LIABLE TO ANY PARTY FOR DIRECT, INDIRECT, SPECIAL, INCIDENTAL, OR CONSEQUENTIAL DAMAGES, INCLUDING LOST PROFITS, ARISING OUT OF THE USE OF THIS CODE, EVEN IF THE AUTHORS HAS BEEN ADVISED OF THE POSSIBILITY OF SUCH DAMAGE. THE AUTHORS SPECIFICALLY DISCLAIMS ANY WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE. FOR RESEARCH PURPOSE ONLY. THIS CODE IS PROVIDED ON A “AS IS” BASIS AND THE AUTHORS HAVE NO OBLIGATION TO PROVIDE MAINTENANCE, SUPPORT, UPDATES, ENHANCEMENTS, OR MODIFICATIONS.
Download
Please follow this link (will open a new page) and fill out the form. An e-mail will be sent to you with a personal download link.